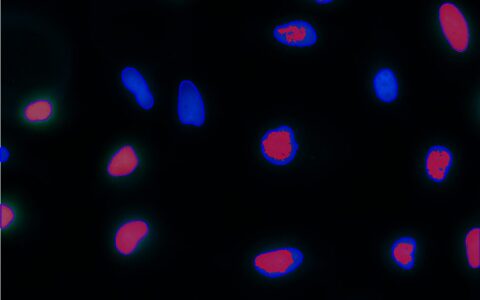
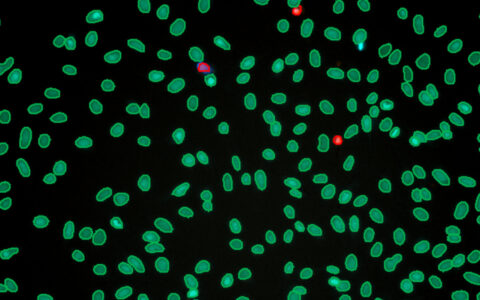
Cell Count (Label-Free)
Cell culture is a fundamental practice in the life sciences, with cell counting being essential at various stages of most experiments involving cultured cells. This includes determining if cell density is sufficient to start an experiment or assessing the effects of an agonist on cell growth and division.
Traditionally, counting cells has been challenging, often requiring fluorescent staining (Ligasová & Koberna 2019) or complex image processing for unlabelled cells (Lin et al. 2022). Deep learning models now enable accurate segmentation of unlabelled cells or nuclei, simplifying the process. The Image-Pro Cell Count (Label-Free) protocol, combined with these models, allows easy analysis of large datasets, such as multi-well plates, even for users with little to no image analysis experience. These models work effectively with phase contrast, pseudo-phase contrast, or differential interference contrast (DIC) imaging.
Techniques: Brightfield
How it works
Select Channel
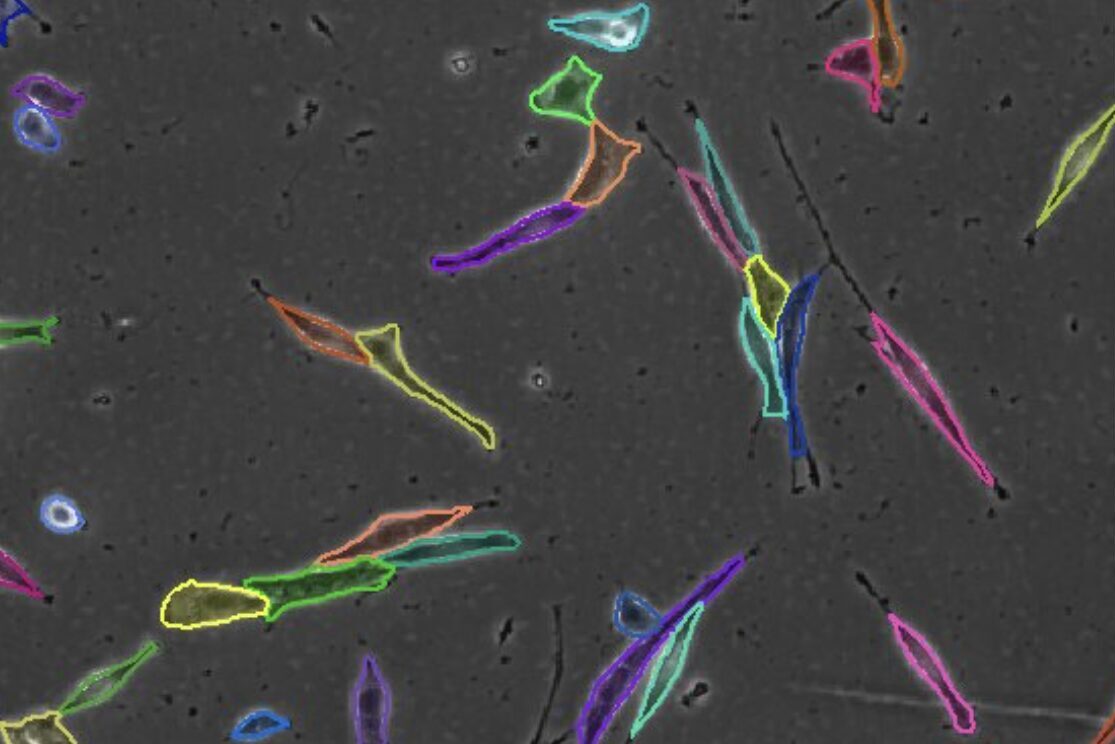
Select the channel that contains unlabeled cells.
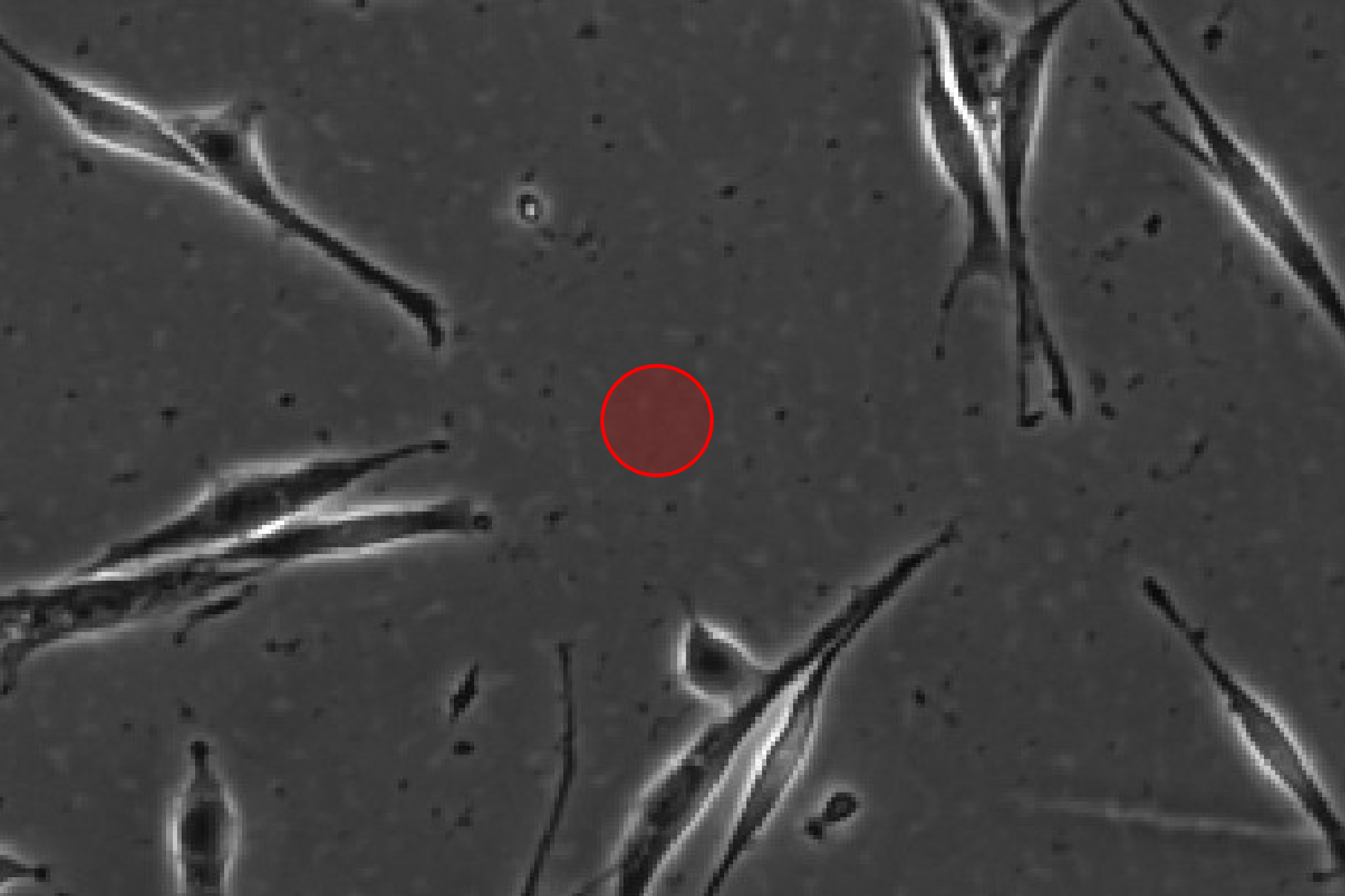
Set Object Diameter
Set the diameter of unlabeled cells.
Find Cells
Find cells or nuclei with a pre-trained deep learning model.
Quantitative results
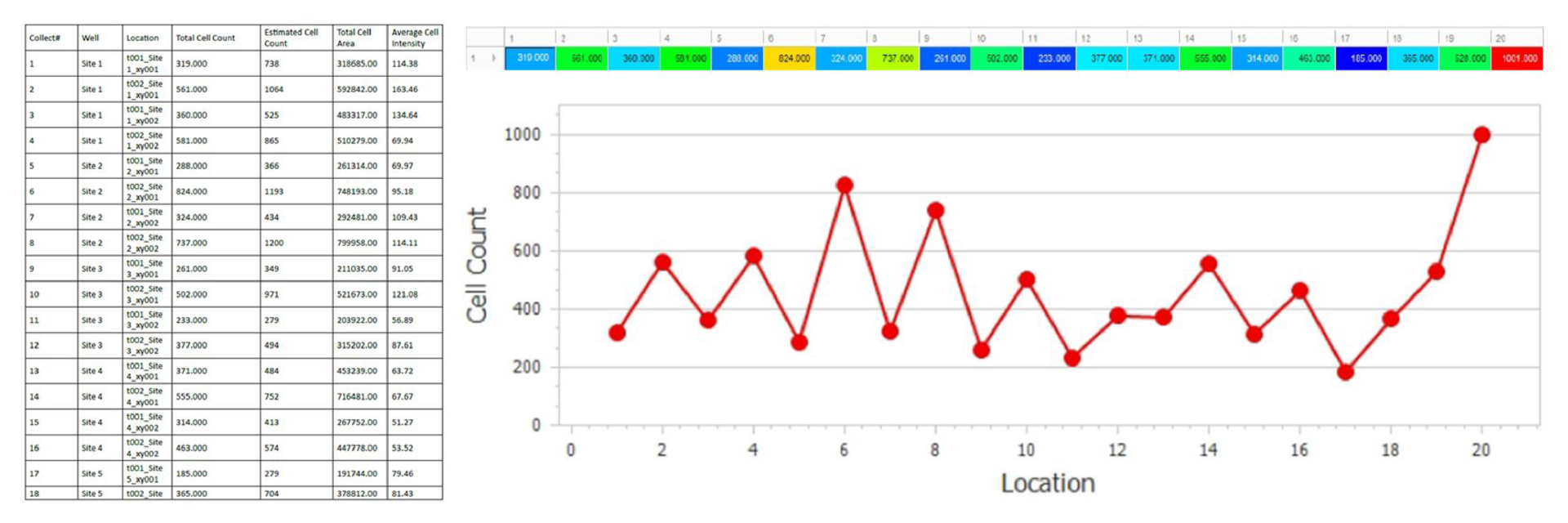
Automatically generate tables, heat maps, charts and even complex bespoke reports.
Measurement parameters supported
- • Total Cell Count
- • Total Cell Area
- • Average Cell Area
- • Custom user-defined measurements
Solution requirements
Required Modules
Base
2D Automated Analysis
Cell Biology Protocol Collection
Cell Count Protocol
AI Deep Learning
Life Science Models
Label Free Cells Model
Recommended Package