Nuclei & Chromatin (TEM)
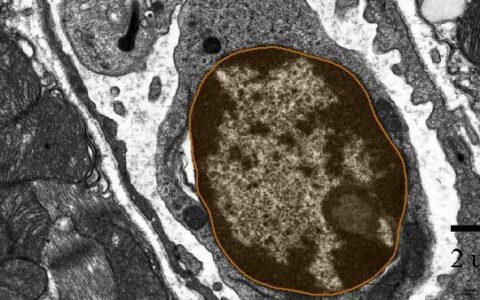
Nuclei are double-membrane-bound organelles that house chromosomes, defining eukaryotic cells. While familiar to all cell biology students through textbooks and electron micrographs, they remain challenging to automatically segment in electron micrographs due to their similar staining characteristics and vast diversity of shapes.
Studies of organelle ultrastructure often rely on the slow and difficult process of manual segmentation (Perez et al., 2014). The Image-Pro pre-trained TEM Nuclei deep learning model overcomes this challenge, working either independently or with the Nuclei and Chromatin (EM) protocol. These tools simplify the analysis of nuclei and chromatin in large volumes of TEM data, requiring little to no image analysis experience.
Techniques: FIB-SEM, TEM
How it works
Select Image
Select your TEM image and open the Nuclei and Chromatin (EM) protocol.
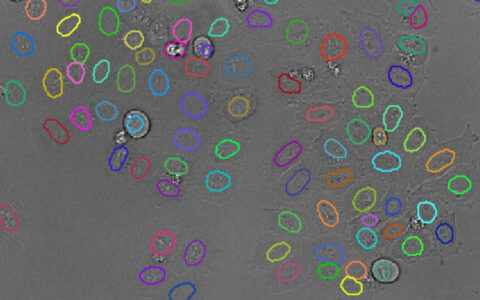
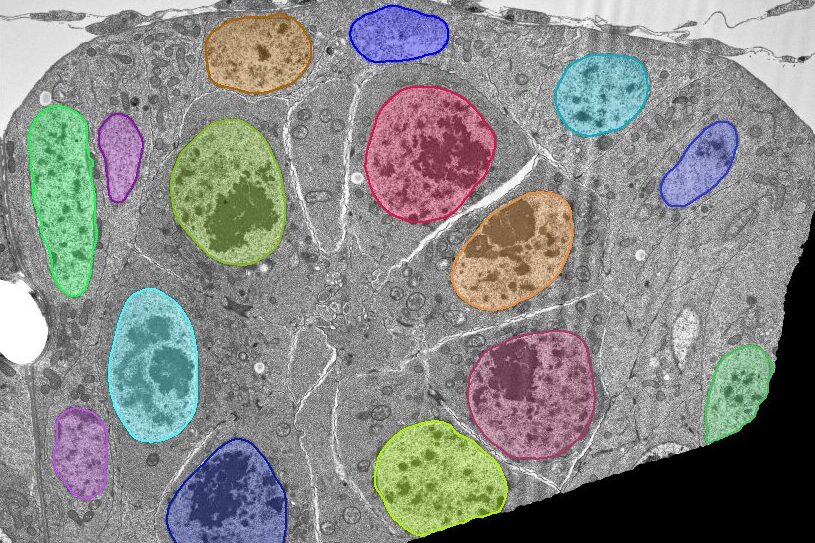
Find Nuclei
Find nuclei with a pre-trained deep learning model.
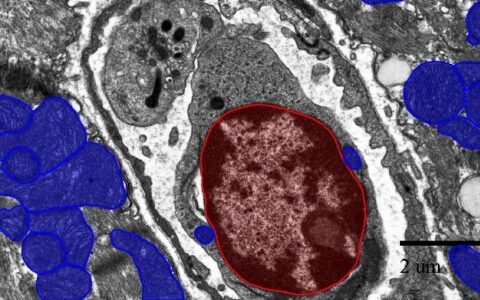
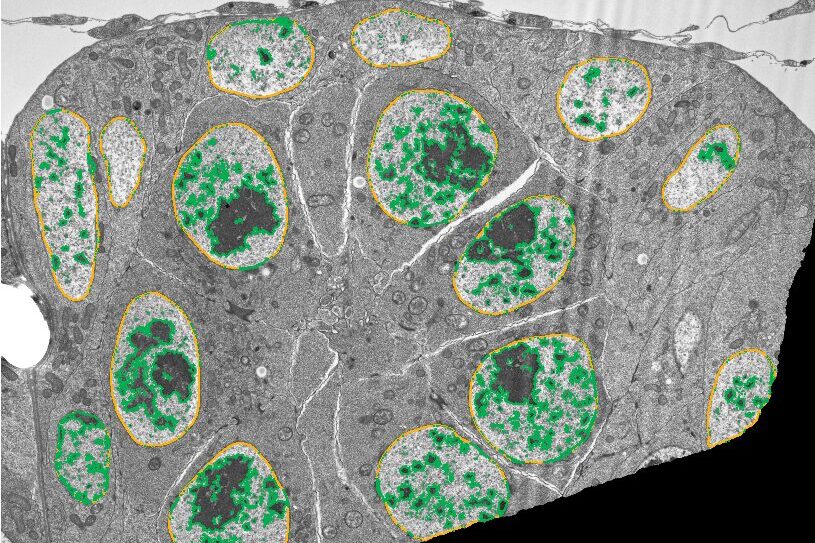
Find Chromatin
Analyze the contents of nuclei, such as the area occupied by darkly staining chromatin.
Quantitative results
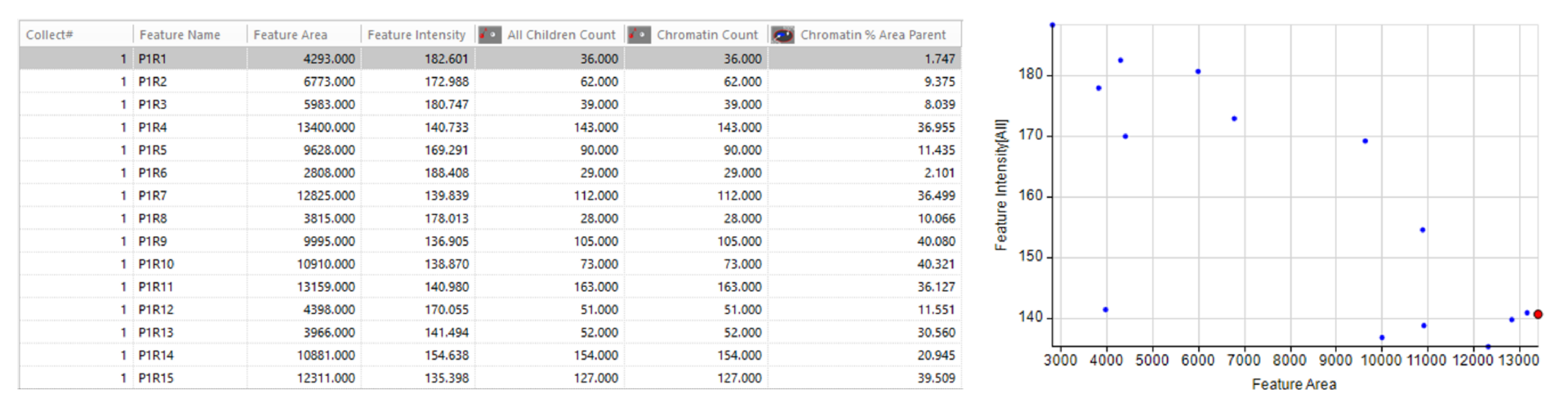
Automatically generate tables, heat maps, charts and even complex bespoke reports.
Measurement parameters supported
- • Nuclei count
- • Percentage of nuclei area occupied by chromatin (mean)
- • Nuclei intensity (mean)
- • Objects per nucleus
- • Custom user-defined measurements
Solution requirements
Required Modules
Base
2D Automated Analysis
Cell Biology EM Protocol Collection
Nuclei and Chromatin (EM) Protocol
AI Deep Learning
Life Science Models
TEM Nuclei Model
Recommended Package