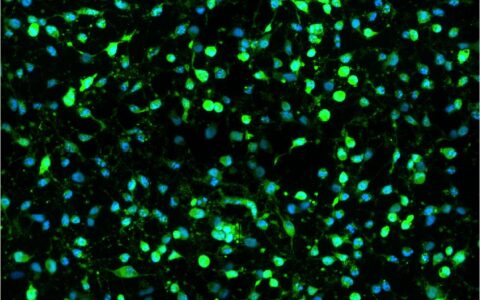
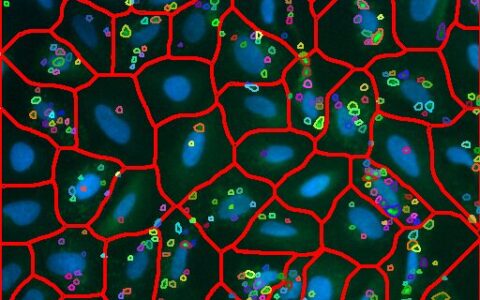
Live/Dead for Cell Viability
Cell viability assays measure the proportion of live and healthy cells in a culture, often used to screen cell populations' response to drugs or chemical agents (Kamiloglu et al., 2020). Cell viability can be assessed by staining live cells with dyes such as Calcein-AM (which is converted to a fluorescent form in living cells) or by detecting dead cells with stains like Propidium Iodide (which is excluded from healthy cells but stains the DNA of dying or dead cells). These assays can even measure both live and dead cells simultaneously.
The Image-Pro Live/Dead Cell Viability protocol is compatible with different combinations, including a) live and total cells, b) dead and total cells, or c) live, dead, and total cells. Pre-configured versions of the protocol are available for these combinations, making it easy to analyze large datasets in complex formats, such as multi-well plates, with minimal image analysis experience.
Techniques: Fluorescence
How it works
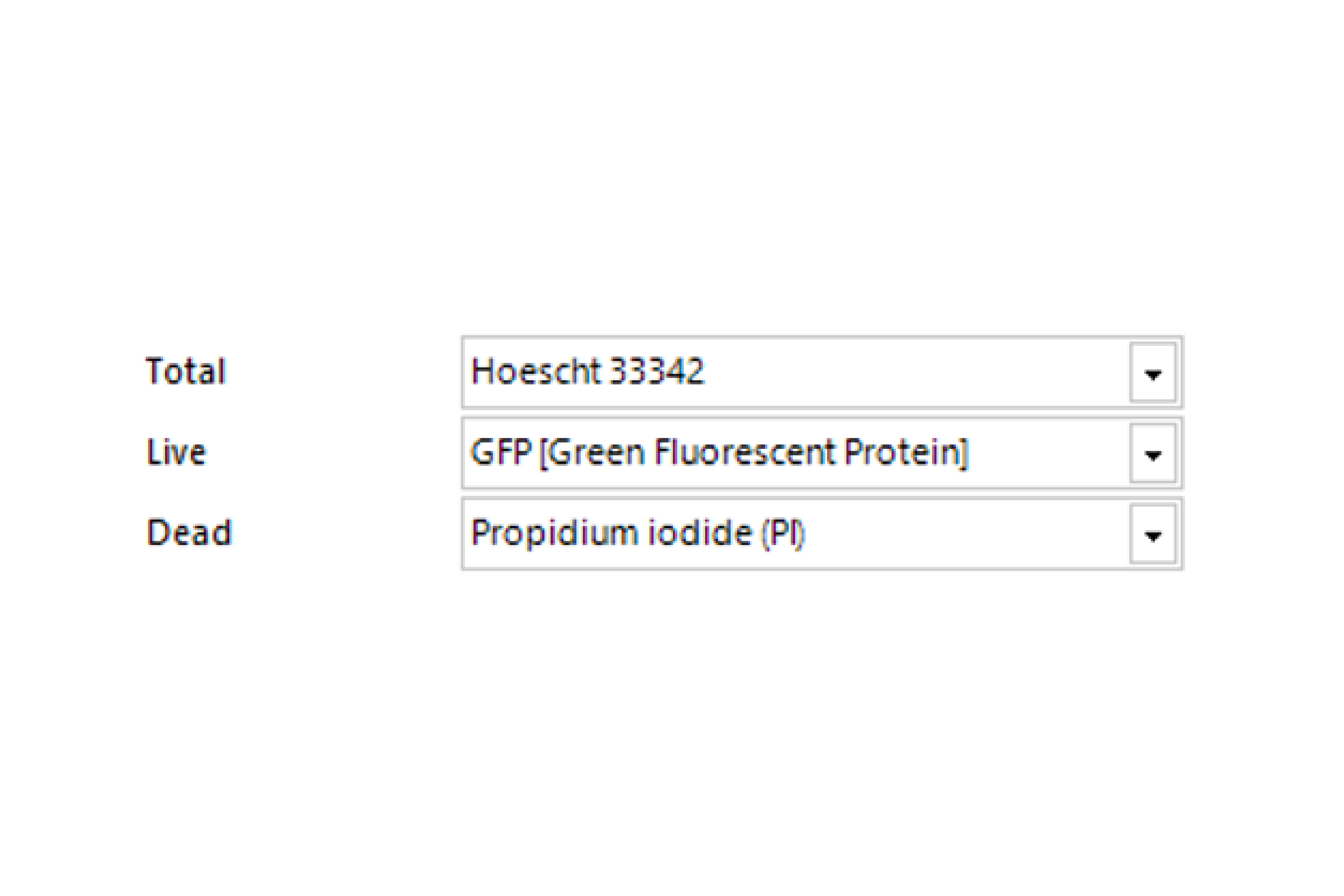
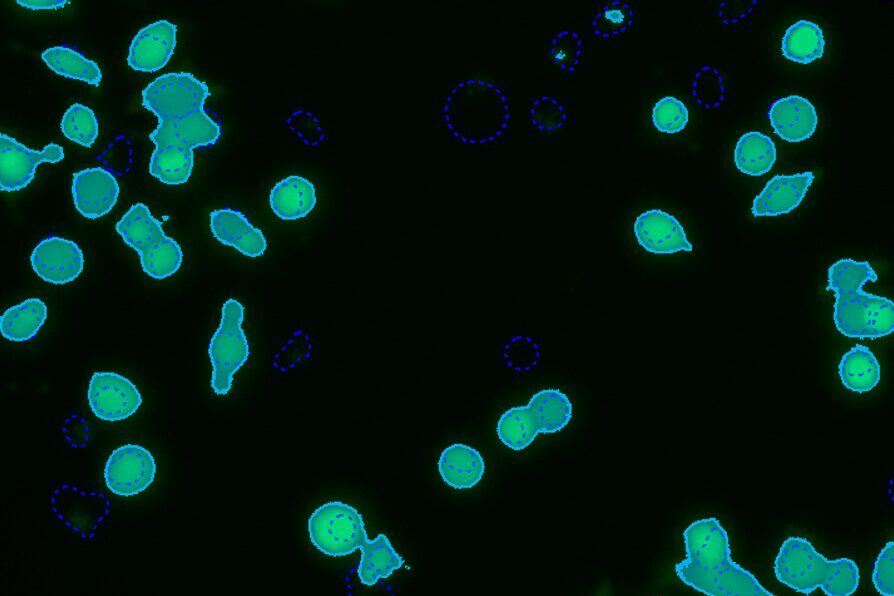
Select Channel
Select the channels that contain total cells, labeled live cells, and labeled dead cells.
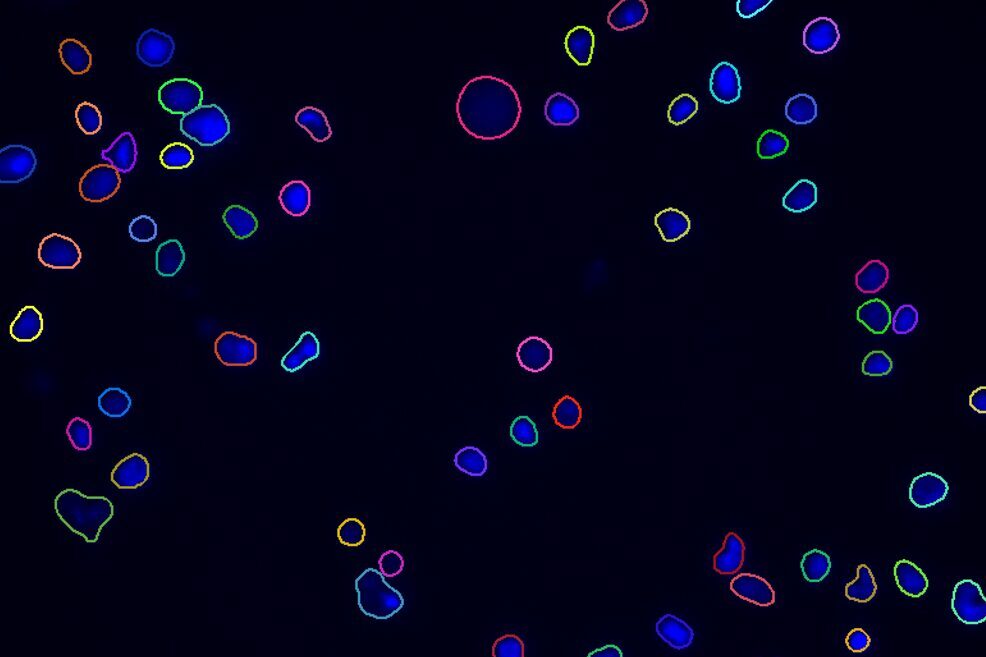
Find Cells or Nuclei
Find cells or nuclei with a pre-trained deep learning model, machine learning, or threshold segmentation.
Find Live/Dead Cells
Identify cells that are labeled as ‘live’ or 'dead' with either threshold segmentation or machine learning segmentation.
Quantitative results
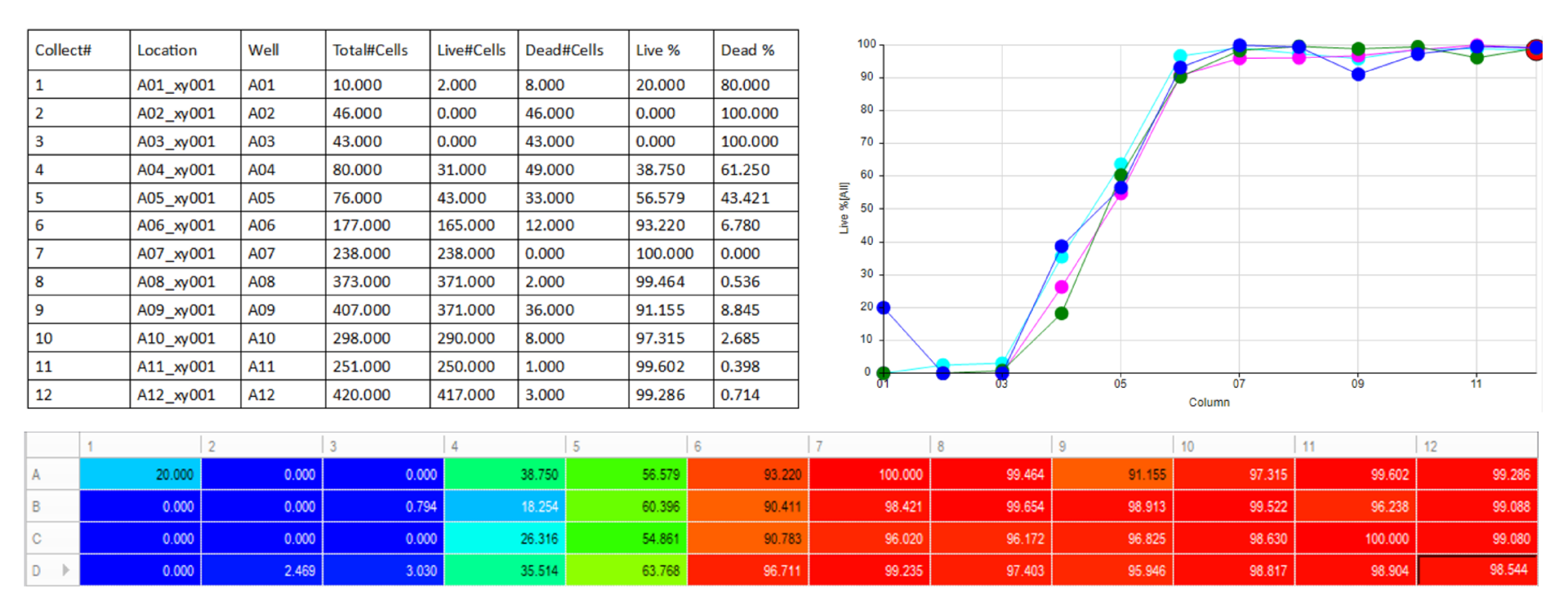
Automatically generate tables, heat maps, charts and even complex bespoke reports.
Measurement parameters supported
- • Live Cell Count
- • Dead Cell Count
- • Total Cells
- • Live Percentage
- • Dead Percentage
- • Custom user-defined measurements
Solution requirements
Required Modules
Base
2D Automated Analysis
Cell Biology Protocol Collection
Live/Dead Cell Viability Protocol
AI Deep Learning
Life Science Models
Fluorescent Cells Model
Recommended Package
Literature spotlight
- Parsons, D., Meredith, K., Rowlands, V. J., Short, D., Metcalf, D. G., & Bowler, P. G. (2016). Enhanced performance and mode of action of a novel antibiofilm Hydrofiber® wound dressing. BioMed Research International, 2016(1), 7616471.
- Kesler Shvero, D., Zaltsman, N., Weiss, E. I., Polak, D., Hazan, R., & Beyth, N. (2016). Lethal bacterial trap: cationic surface for endodontic sealing. Journal of Biomedical Materials Research Part A, 104(2), 427-434.
- Xavier, M. V., Macedo, M. F., Benatti, A. C. B., Jardini, A. L., Rodrigues, A. A., Lopes, M. S., ... & Kharmandayan, P. (2016). PLLA synthesis and nanofibers production: viability by human mesenchymal stem cell from adipose tissue. Procedia CIRP, 49, 213-221.4