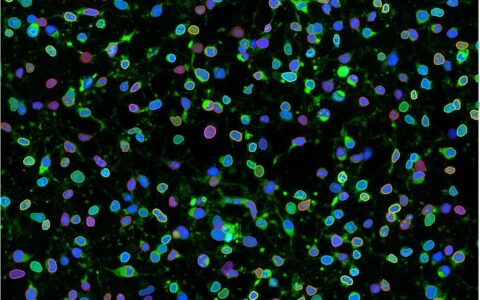
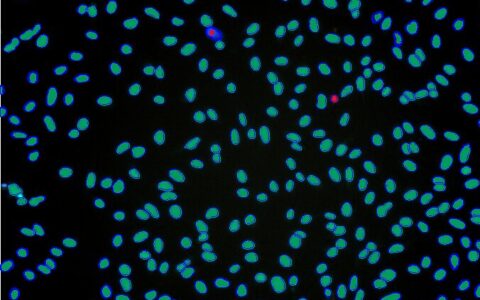
Cell Autophagy
Autophagy is a conserved cellular recycling process in eukaryotes, responsible for removing unnecessary or damaged components through a lysosome-dependent mechanism. This process is critical for cell survival and maintenance, and its dysfunction is associated with various human diseases (Parzych et al., 2014).
Fluorescent dye-based assays are commonly used to label autophagosomes for analysis (Ding and Hong, 2020). The Image-Pro Autophagy protocol streamlines the analysis of these assays, supporting large-scale data processing in complex formats like multi-well plates. Its intuitive design allows even users with minimal image analysis experience to obtain accurate and reproducible results efficiently.
Techniques: Fluorescence
How it works
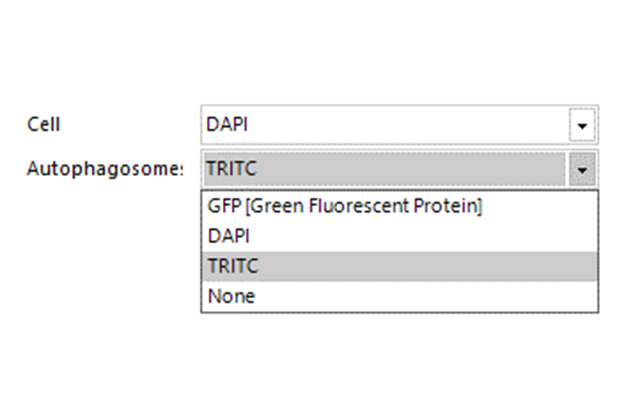
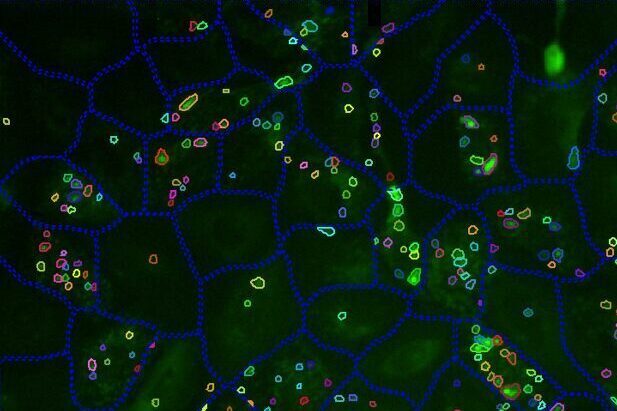
Select Channel
Select the channels that contain labeled cells (cellular or nuclear dye) and labeled autophagosomes.
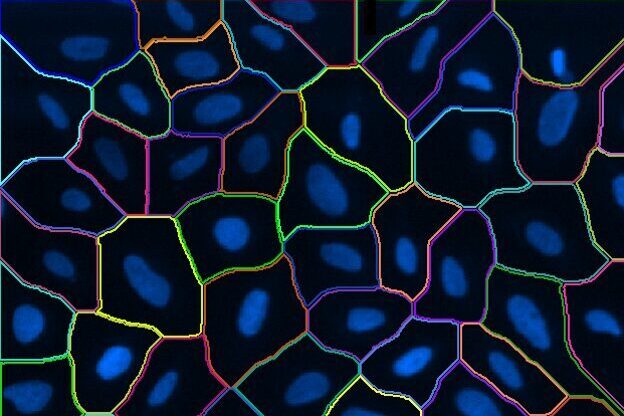
Find Cells
Find cells or nuclei with a pre-trained deep learning model, machine learning, or threshold segmentation.
Find Autophagosomes
Find autophagosomes with a pre-trained deep learning model, machine learning, or threshold segmentation.
Quantitative results
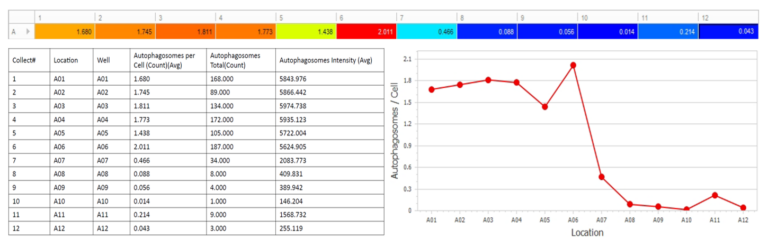
Automatically generate tables, heat maps, charts and even complex bespoke reports.
Measurement parameters supported
- • Autophagosomes Per Cell
- • Mean Autophagosome Diameter
- • Mean Autophagosome Intensity
- • Mean Autophagosome Area
- • Autophagosome Count
- • Custom user-defined measurements
Solution requirements
Required Modules
Base
2D Automated Analysis
Cell Biology Plus Protocol Collection
Cell Autophagy Procol
AI Deep Learning
Life Science Models
Fluorescent Nuclei Model
Recommended Package
Literature spotlight
- Ji, L., Wang, Z.H., Zhang, Y.H., Zhou, Y., Tang, D.S., Yan, C.S., Ma, J.M., Fang, K., Gao, L., Ren, N.S. and Cheng, L., 2022. ATG7-enhanced impaired autophagy exacerbates acute pancreatitis by promoting regulated necrosis via the miR-30b-5p/CAMKII pathway. Cell Death & Disease, 13(3), p.211.
- Wu, Dongle, Xuan Sun, Yiwei Zhao, Yuanbo Liu, Ziqi Gan, Zhen Zhang, Xin Chen, and Yang Cao. "Strontium ranelate inhibits osteoclastogenesis through NF-κB-pathway-dependent autophagy." Bioengineering 10, no. 3 (2023): 365.
- Zou, Y., Zhang, X., Chen, X.Y., Ma, X.F., Feng, X.Y., Sun, Y., Ma, T., Ma, Q.H., Zhao, X.D. and Xu, D.E., (2024) Contactin-Associated protein1 Regulates Autophagy by Modulating the PI3K/AKT/mTOR Signaling Pathway and ATG4B Levels in Vitro and in Vivo. Molecular Neurobiology, pp.1-17.