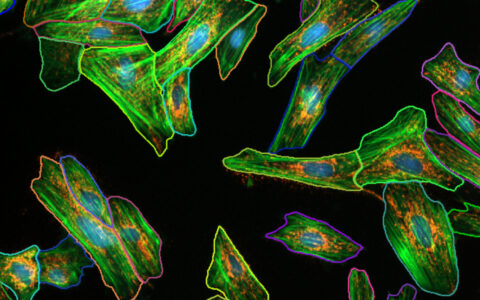
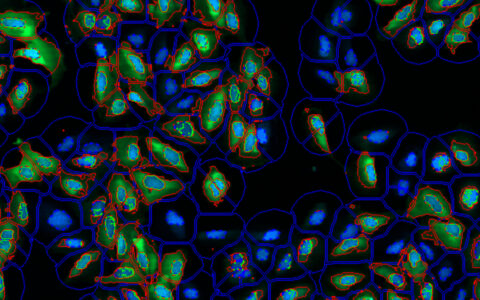
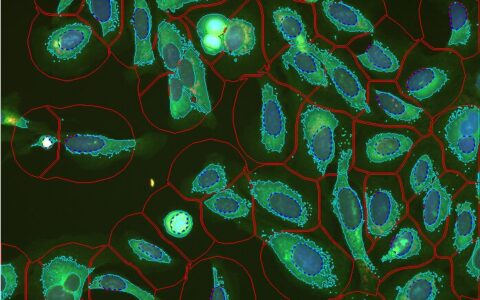
Neurite Outgrowth
Neurons are essential components of nervous tissue, and studying neurodegenerative disorders like Alzheimer’s and Parkinson’s diseases presents significant challenges (Wang et al., 2010). In-vitro studies of neuronal development in both normal and disease states require complex image analysis to measure key metrics like neurite growth, branching, and termini, which demand advanced computational tools (Meijering, 2010).
The Image-Pro Neurite Outgrowth protocol simplifies the analysis of large, complex datasets, such as multi-well plates, with minimal image analysis experience. This makes it easier for researchers to efficiently analyze neuronal development and support therapeutic advancements.
Techniques: Brightfield, Fluorescence
How it works

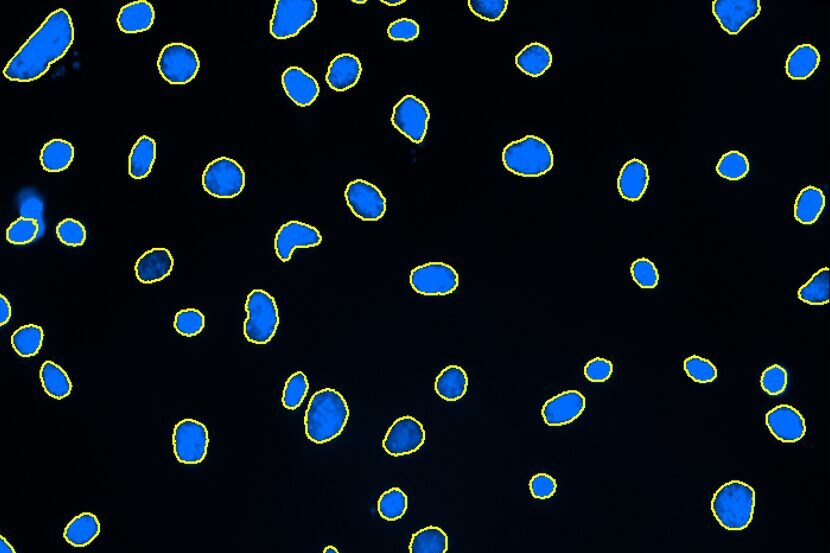
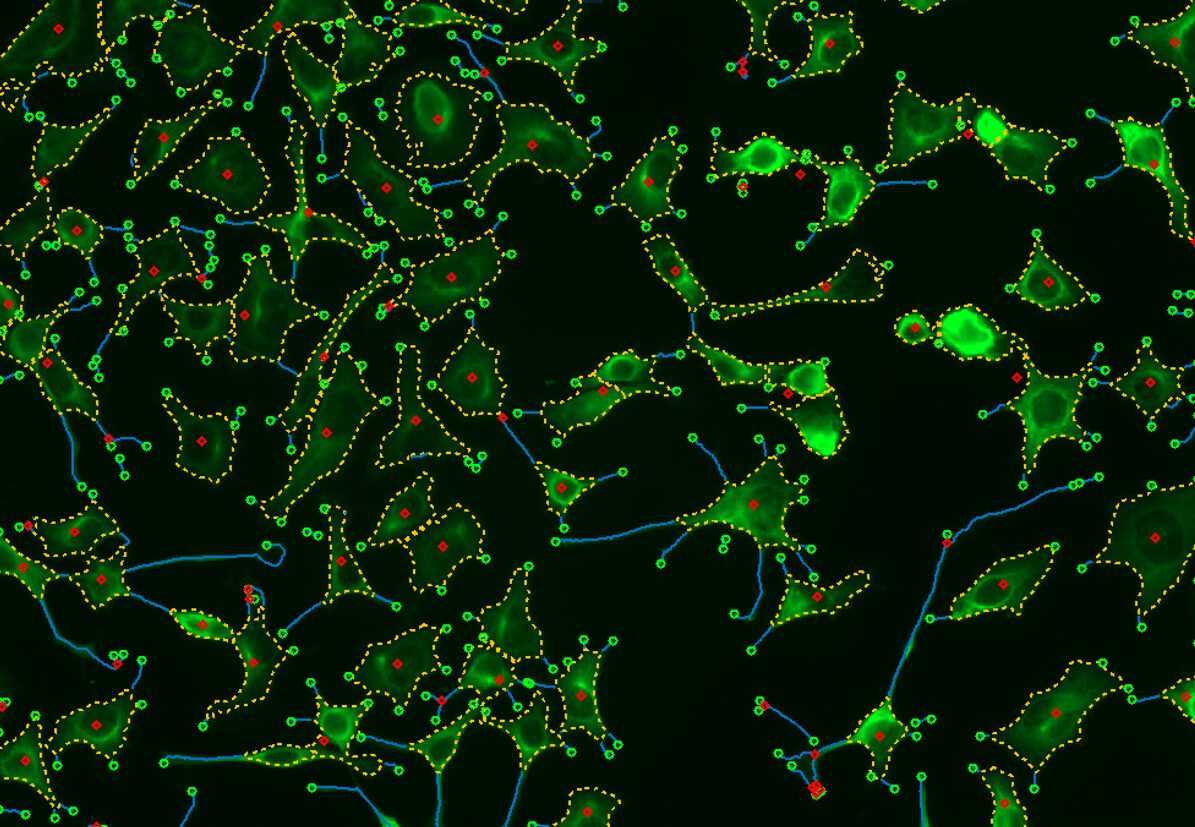
Select Channel
Select the channels that contain labeled nuclei and neurites.
Find Nuclei
Find nuclei with a pre-trained deep learning model, machine learning, or threshold segmentation.
Find Cell Bodies & Neurites
Find cell bodies with simple sliders. Find neurites with either a pre-trained deep learning model or simple sliders.
Quantitative results
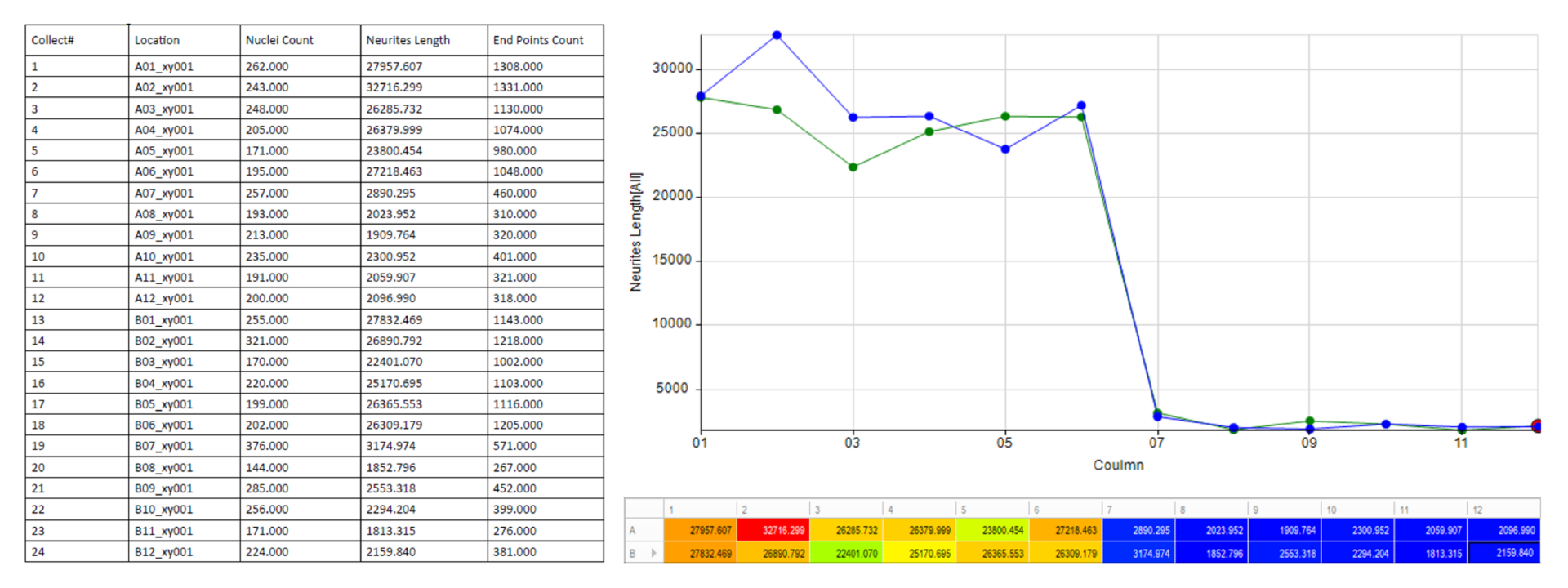
Automatically generate tables, heat maps, charts and even complex bespoke reports.
Measurement parameters supported
- • Total Neurite Length
- • End Point Count
- • Branch Count
- • Nuclei Count
- • Custom user-defined measurements
Solution requirements
Required Modules
Base
2D Automated Analysis
Cell Biology Plus Protocol Collection
Neurite Outgrowth Protocol
AI Deep Learning
Life Science Models
Fluorescent Dendrites Model
Recommended Package