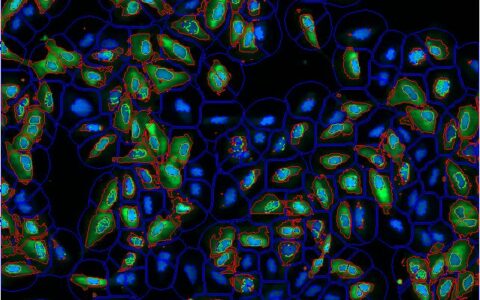
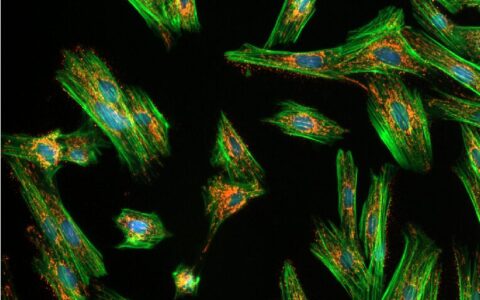
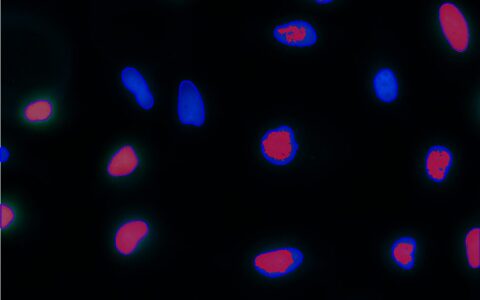
Colocalization
A common question among cell biologists is whether a pair of fluorescently labeled proteins interact with one another. This can often be deduced by examining micrographs—if the proteins are localized in the same areas, there is a chance they interact. If their intensity distributions correlate, with both becoming brighter or darker in the same regions, the likelihood of interaction is increased.
The Image-Pro Colocalization protocol calculates Pearson's Correlation Coefficient (PCC) between two fluorescence channels, quantifying the degree of overlap between them. It also calculates the proportion of intensities in each channel to which the PCC applies and summarizes the results in an easy-to-understand sentence. This protocol allows for the analysis of large datasets in formats like multi-well plates, requiring little to no image analysis experience.
Techniques: Fluorescence
How it works
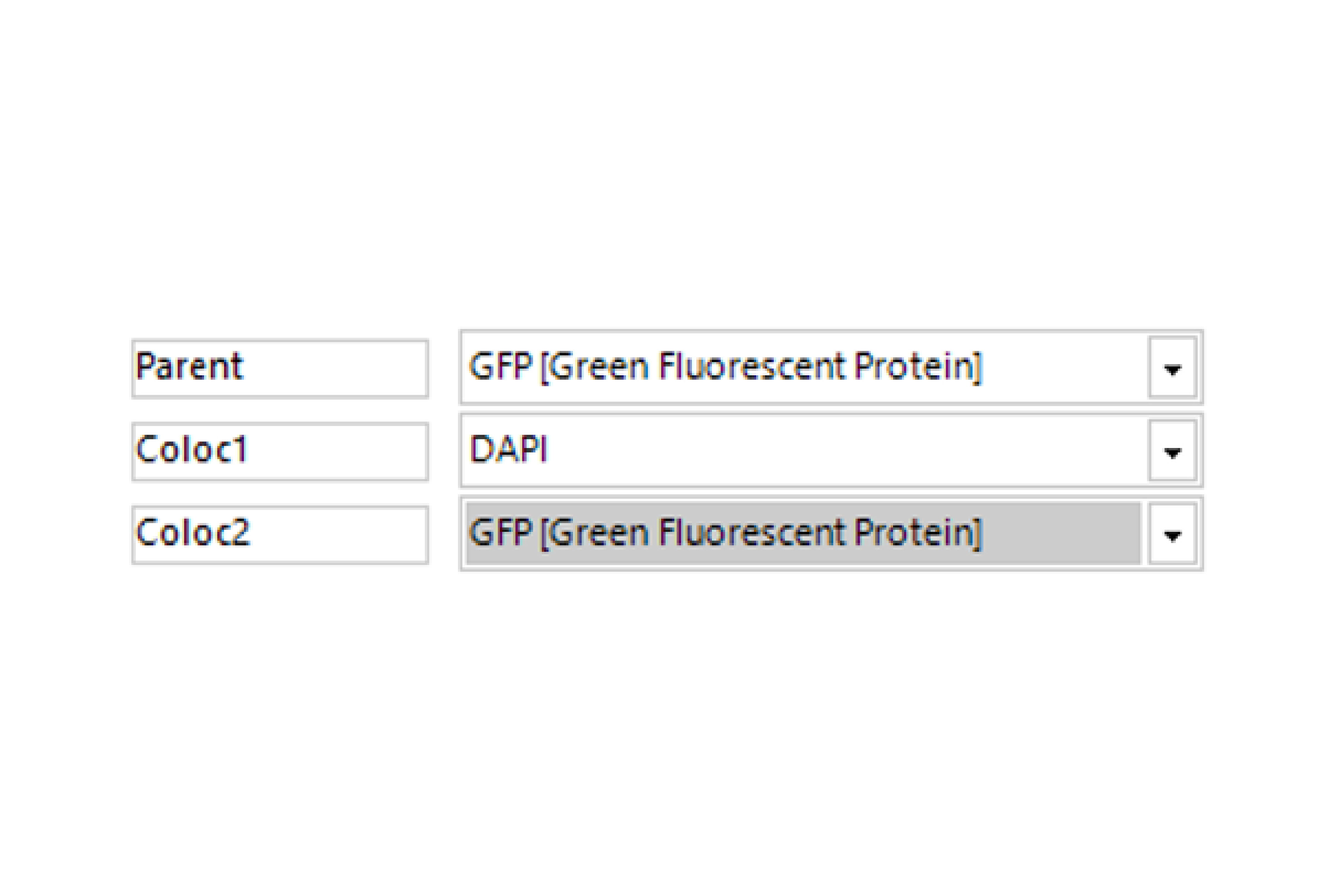
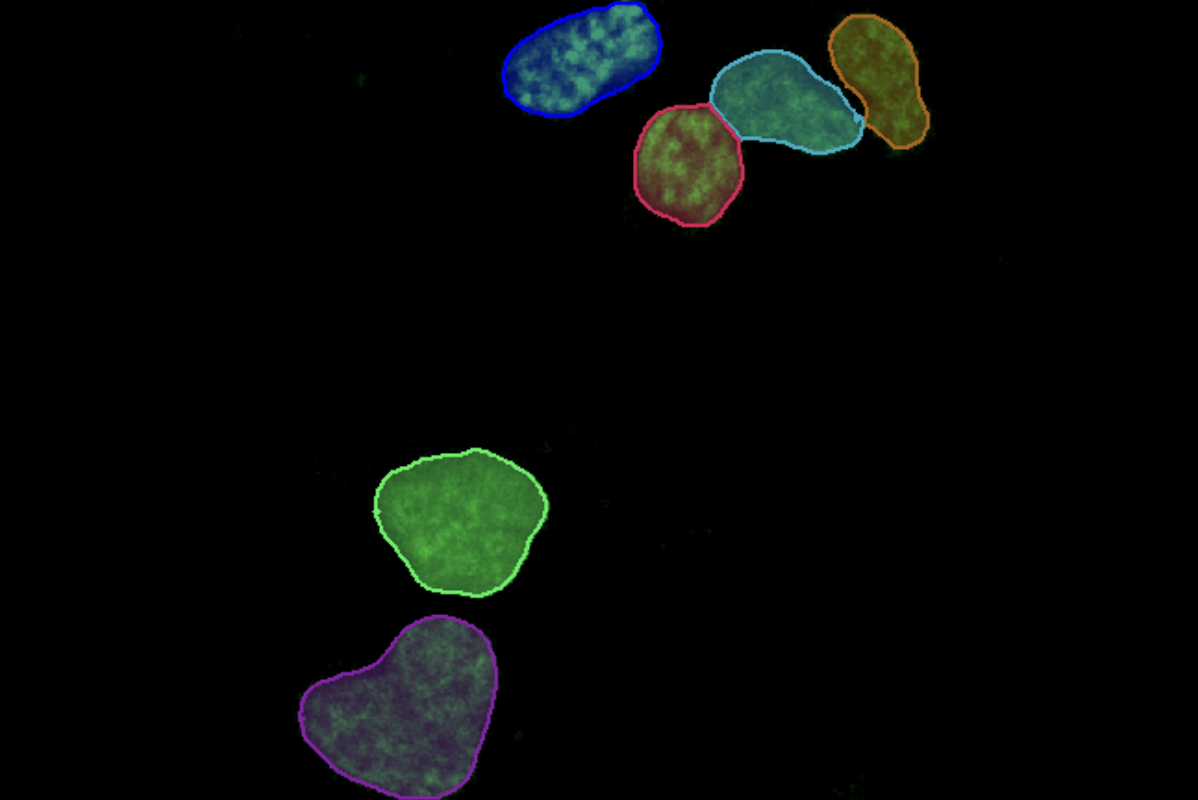
Select Channel
Select the channels that contains labeled parent objects (optional) and the colocalization analysis channels.
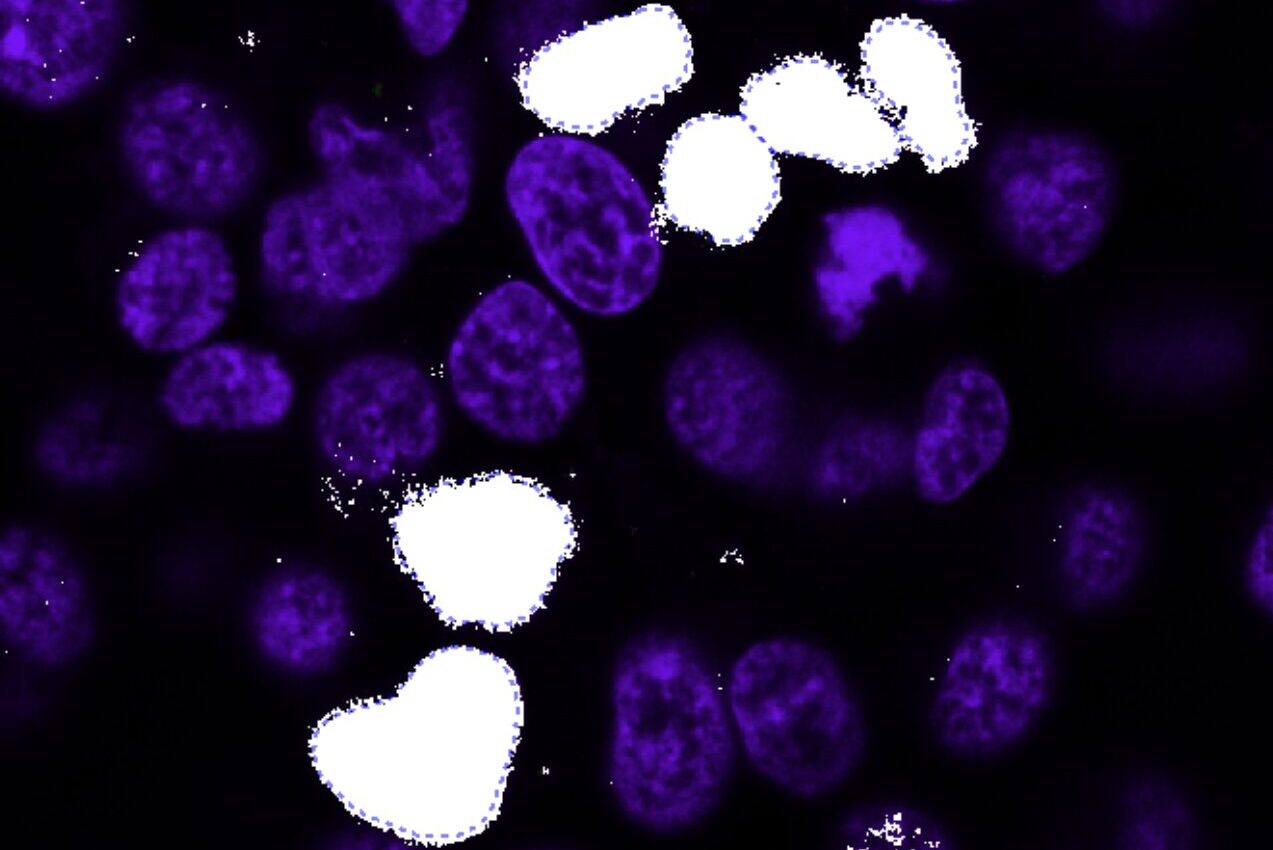
Find Parent Objects
Find parent objects with a pre-trained deep learning model, machine learning, or threshold segmentation.
Calculate Thresholds
Automatically calculate the thresholds for the colocalization analysis channels.
Quantitative results
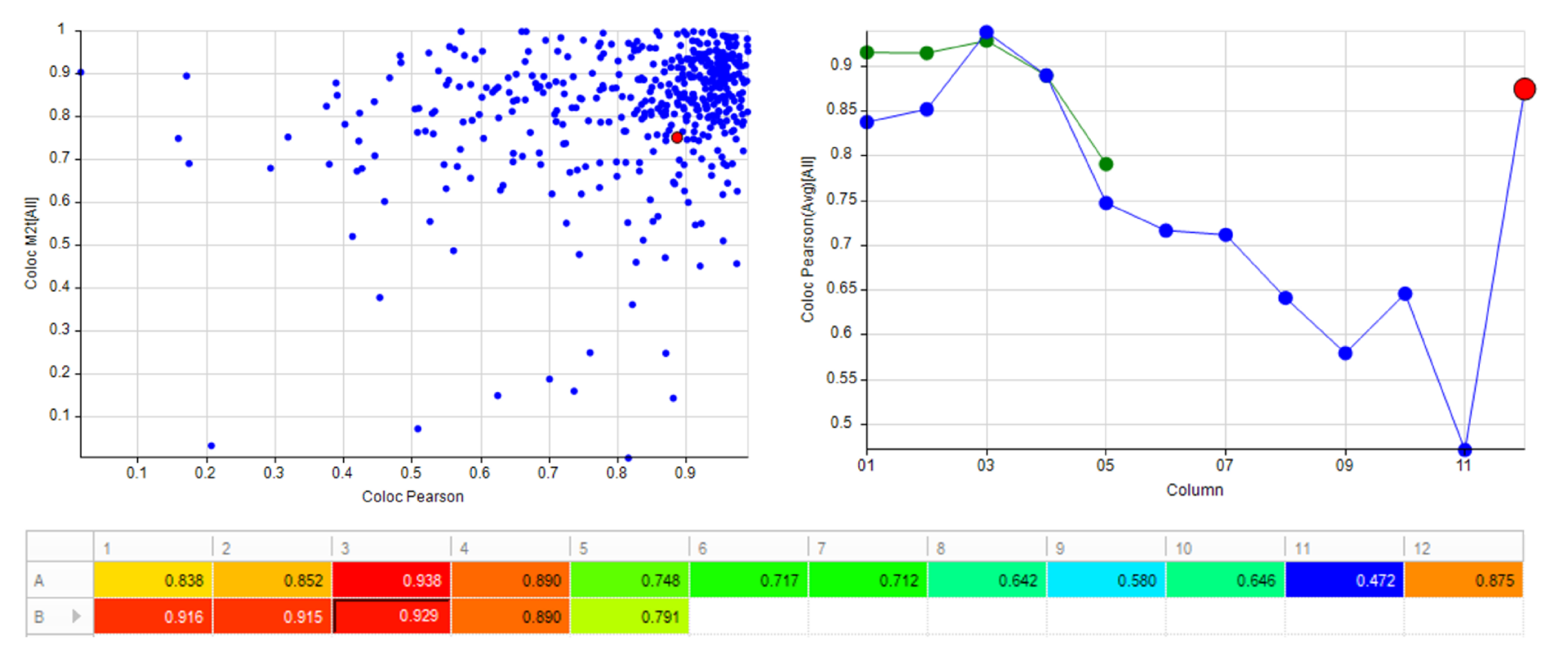
Automatically generate tables, heat maps, charts and even complex bespoke reports.
Measurement parameters supported
- • Pearsons Correlation Coefficient
- • Manders Overlap Coefficients (M1t and M2t)
- • A simple interpretive sentence (E.g. “There is a mean positive correlation of 0.874. This applies to a mean of 100% of Image 1 colocalized object intensities, and 91.3% of Image 2 colocalized object intensities.”)
- • Custom user-defined measurements
Solution requirements
Required Modules
Base
2D Automated Analysis
Cell Biology Protocol Collection
Colocalization Protocol
AI Deep Learning
Life Science Models
Fluorescent Cells Model
Recommended Package
Literature spotlight
- Guo, Q. Q., Gao, J., Wang, X. W., Yin, X. L., Zhang, S. C., Li, X., ... & Zhang, Q. Y. (2021). RNA-binding protein MSI2 binds to miR-301a-3p and facilitates its distribution in mitochondria of endothelial cells. Frontiers in Molecular Biosciences, 7, 609828.
- Toro-Fernández, L. F., Zuluaga-Monares, J. C., Saldarriaga-Cartagena, A. M., Cardona-Gómez, G. P., & Posada-Duque, R. (2021). Targeting CDK5 in astrocytes promotes calcium homeostasis under excitotoxic conditions. Frontiers in Cellular Neuroscience, 15, 643717.
- Wen, Q. X., Luo, B., Xie, X. Y., Zhou, G. F., Chen, J., Song, L., ... & Chen, G. J. (2023). AP2S1 regulates APP degradation through late endosome–lysosome fusion in cells and APP/PS1 mice. Traffic, 24(1), 20-33.